1 + 1[1] 2In this Chapter we’ll cover how to:
We provide screenshot of RStudio but everything is really similar when using VSCode.
Before we continue, here are a few things to bear in mind as you work through this Chapter:
R is case sensitive i.e. A is not the same as a and anova is not the same as Anova.
Anything that follows a # symbol is interpreted as a comment and ignored by R. Comments should be used liberally throughout your code for both your own information and also to help your collaborators. Writing comments is a bit of an art and something that you will become more adept at as your experience grows.
In R, commands are generally separated by a new line. You can also use a semicolon ; to separate your commands but we strongly recommend to avoid using it.
If a continuation prompt + appears in the console after you execute your code this means that you haven’t completed your code correctly. This often happens if you forget to close a bracket and is especially common when nested brackets are used ((((some command))). Just finish the command on the new line and fix the typo or hit escape on your keyboard (see point below) and fix.
In general, R is fairly tolerant of extra spaces inserted into your code, in fact using spaces is actively encouraged. However, spaces should not be inserted into operators i.e. <- should not read < - (note the space). See the style guide for advice on where to place spaces to make your code more readable.
If your console ‘hangs’ and becomes unresponsive after running a command you can often get yourself out of trouble by pressing the escape key (esc) on your keyboard or clicking on the stop icon in the top right of your console. This will terminate most current operations.
In Chapter 1, we learned about the R Console and creating scripts and Projects. We also saw how you write your R code in a script and then source this code into the console to get it to run (if you’ve forgotten how to do this, pop back to the console section (1.2.1.1) to refresh your memory). Writing your code in a script means that you’ll always have a permanent record of everything you’ve done (provided you save your script) and also allows you to make loads of comments to remind your future self what you’ve done. So, while you’re working through this Chapter we suggest that you create a new script (or RStudio Project) to write your code as you follow along.
As we saw in Chapter 1, at a basic level we can use R much as you would use a calculator. We can type an arithmetic expression into our script, then source it into the console and receive a result. For example, if we type the expression 1 + 1 and then source this line of code we get the answer 2 (😃!)
1 + 1[1] 2The [1] in front of the result tells you that the observation number at the beginning of the line is the first observation. This is not much help in this example, but can be quite useful when printing results with multiple lines (we’ll see an example below). The other obvious arithmetic operators are -, *, / for subtraction, multiplication and division respectively. Matrix multiplication operator is %*%. R follows the usual mathematical convention of order of operations. For example, the expression 2 + 3 * 4 is interpreted to have the value 2 + (3 * 4) = 14, not (2 + 3) * 4 = 20. There are a huge range of mathematical functions in R, some of the most useful include; log(), log10(), exp(), sqrt().
log(1) # logarithm to base e[1] 0log10(1) # logarithm to base 10[1] 0exp(1) # natural antilog[1] 2.718282sqrt(4) # square root[1] 24^2 # 4 to the power of 2[1] 16pi # not a function but useful[1] 3.141593It’s important to realize that when you run code as we’ve done above, the result of the code (or value) is only displayed in the console. Whilst this can sometimes be useful it is usually much more practical to store the value(s) in aN object.
At the heart of almost everything you will do (or ever likely to do) in R is the concept that everything in R is an object. These objects can be almost anything, from a single number or character string (like a word) to highly complex structures like the output of a plot, a summary of your statistical analysis or a set of R commands that perform a specific task. Understanding how you create objects and assign values to objects is key to understanding R.
To create an object we simply give the object a name. We can then assign a value to this object using the assignment operator <- (sometimes called the gets operator). The assignment operator is a composite symbol comprised of a ‘less than’ symbol < and a hyphen - .
my_obj <- 32In the code above, we created an object called my_obj and assigned it a value of the number 32 using the assignment operator (in our head we always read this as ‘my_obj is 32’). You can also use = instead of <- to assign values but this is bad practice since it can lead to confusion later on when programming in R (see Chapter 5) and we would discourage you from using this notation.
To view the value of the object you simply type the name of the object
my_obj[1] 32Now that we’ve created this object, R knows all about it and will keep track of it during this current R session. All of the objects you create will be stored in the current workspace and you can view all the objects in your workspace in RStudio by clicking on the ‘Environment’ tab in the top right hand pane.
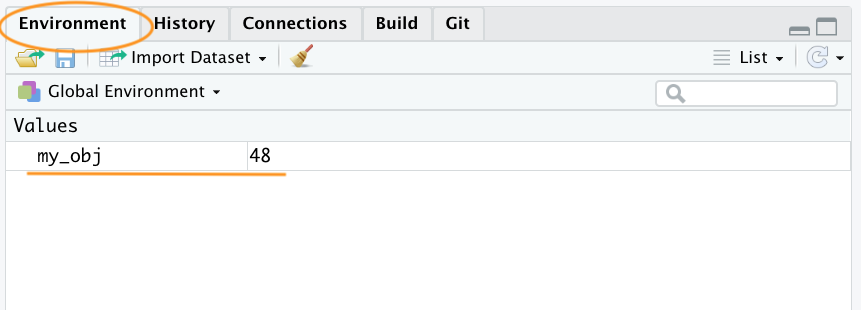
If you click on the down arrow on the ‘List’ icon in the same pane and change to ‘Grid’ view RStudio will show you a summary of the objects including the type (numeric - it’s a number), the length (only one value in this object), its ‘physical’ size and its value (48 in this case). In VSCode, go on the R extension pane, and you can obtain the same information.
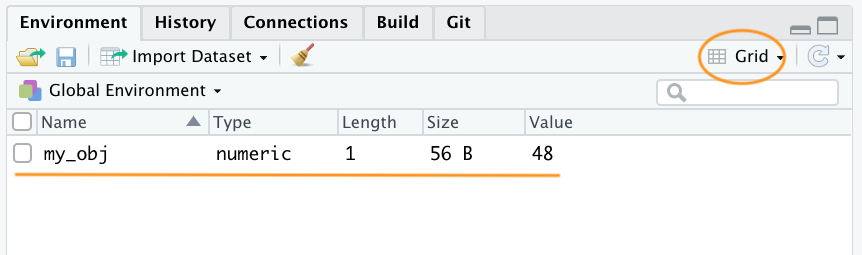
There are many different types of values that you can assign to an object. For example
my_obj2 <- "R is cool"Here we have created an object called my_obj2 and assigned it a value of R is cool which is a character string. Notice that we have enclosed the string in quotes. If you forget to use the quotes you will receive an error message.
Our workspace now contains both objects we’ve created so far with my_obj2 listed as type character.
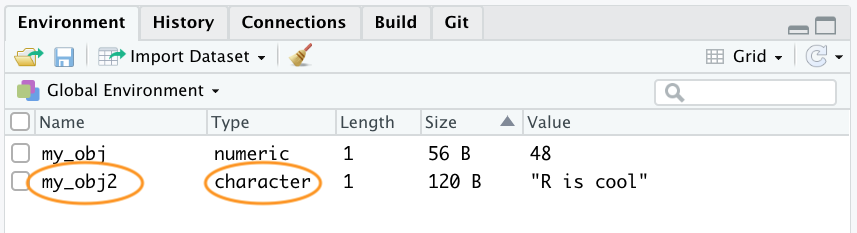
To change the value of an existing object we simply reassign a new value to it. For example, to change the value of my_obj2 from "R is cool" to the number 1024
my_obj2 <- 1024Notice that the Type has changed to numeric and the value has changed to 1024 in the environment
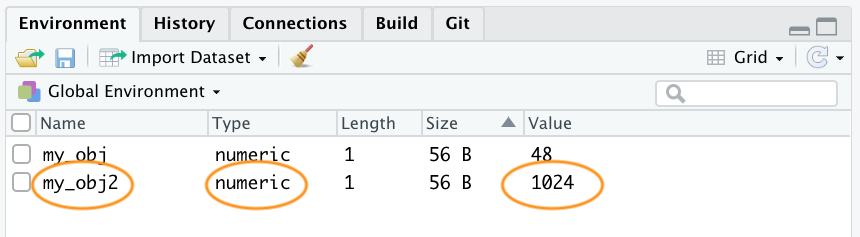
Once we have created a few objects, we can do stuff with our objects. For example, the following code creates a new object my_obj3 and assigns it the value of my_obj added to my_obj2 which is 1072 (48 + 1024 = 1072).
my_obj3 <- my_obj + my_obj2
my_obj3[1] 1056Notice that to display the value of my_obj3 we also need to write the object’s name. The above code works because the values of both my_obj and my_obj2 are numeric (i.e. a number). If you try to do this with objects with character values (character class) you will receive an error
char_obj <- "hello"
char_obj2 <- "world!"
char_obj3 <- char_obj + char_obj2
# Error in char_obj+char_obj2:non-numeric argument to binary operatorThe error message is essentially telling you that either one or both of the objects char_obj and char_obj2 is not a number and therefore cannot be added together.
When you first start learning R, dealing with errors and warnings can be frustrating as they’re often difficult to understand (what’s an argument? what’s a binary operator?). One way to find out more information about a particular error is to search for a generalised version of the error message. For the above error try searching ‘non-numeric argument to binary operator error + r’ or even ‘common r error messages’.
Another error message that you’ll get quite a lot when you first start using R is Error: object 'XXX' not found. As an example, take a look at the code below
my_obj <- 48
my_obj4 <- my_obj + no_obj
# Error: object 'no_obj' not foundR returns an error message because we haven’t created (defined) the object no_obj yet. Another clue that there’s a problem with this code is that, if you check your environment, you’ll see that object my_obj4 has not been created.
Naming your objects is one of the most difficult things you will do in R. Ideally your object names should be kept both short and informative which is not always easy. If you need to create objects with multiple words in their name then use either an underscore or a dot between words or capitalise the different words. We prefer the underscore format and never include uppercase in names (called snake_case)
output_summary <- "my analysis" # recommended#
output.summary <- "my analysis"
outputSummary <- "my analysis"There are also a few limitations when it come to giving objects names. An object name cannot start with a number or a dot followed by a number (i.e. 2my_variable or .2my_variable). You should also avoid using non-alphanumeric characters in your object names (i.e. &, ^, /, ! etc). In addition, make sure you don’t name your objects with reserved words (i.e. TRUE, NA) and it’s never a good idea to give your object the same name as a built-in function. One that crops up more times than we can remember is
data <- read.table("mydatafile", header = TRUE)Yes, data() is a function in R to load or list available data sets from packages.
Up until now we’ve been creating simple objects by directly assigning a single value to an object. It’s very likely that you’ll soon want to progress to creating more complicated objects as your R experience grows and the complexity of your tasks increase. Happily, R has a multitude of functions to help you do this. You can think of a function as an object which contains a series of instructions to perform a specific task. The base installation of R comes with many functions already defined or you can increase the power of R by installing one of the 10,000’s of packages now available. Once you get a bit more experience with using R you may want to define your own functions to perform tasks that are specific to your goals (more about this in Chapter 5).
The first function we will learn about is the c() function. The c() function is short for concatenate and we use it to join together a series of values and store them in a data structure called a vector (more on vectors in Chapter 3).
my_vec <- c(2, 3, 1, 6, 4, 3, 3, 7)In the code above we’ve created an object called my_vec and assigned it a value using the function c(). There are a couple of really important points to note here. Firstly, when you use a function in R, the function name is always followed by a pair of round brackets even if there’s nothing contained between the brackets. Secondly, the argument(s) of a function are placed inside the round brackets and are separated by commas. You can think of an argument as way of customising the use or behaviour of a function. In the example above, the arguments are the numbers we want to concatenate. Finally, one of the tricky things when you first start using R is to know which function to use for a particular task and how to use it. Thankfully each function will always have a help document associated with it which will explain how to use the function (more on this later Section 2.6) and a quick web search will also usually help you out.
To examine the value of our new object we can simply type out the name of the object as we did before
my_vec[1] 2 3 1 6 4 3 3 7Now that we’ve created a vector we can use other functions to do useful stuff with this object. For example, we can calculate the mean, variance, standard deviation and number of elements in our vector by using the mean(), var(), sd() and length() functions
mean(my_vec) # returns the mean of my_vec[1] 3.625var(my_vec) # returns the variance of my_vec[1] 3.982143sd(my_vec) # returns the standard deviation of my_vec[1] 1.995531length(my_vec) # returns the number of elements in my_vec[1] 8If we wanted to use any of these values later on in our analysis we can just assign the resulting value to another object
vec_mean <- mean(my_vec) # returns the mean of my_vec
vec_mean[1] 3.625Sometimes it can be useful to create a vector that contains a regular sequence of values in steps of one. Here we can make use of a shortcut using the : symbol.
my_seq <- 1:10 # create regular sequence
my_seq [1] 1 2 3 4 5 6 7 8 9 10my_seq2 <- 10:1 # in decending order
my_seq2 [1] 10 9 8 7 6 5 4 3 2 1Other useful functions for generating vectors of sequences include the seq() and rep() functions. For example, to generate a sequence from 1 to 5 in steps of 0.5
my_seq2 <- seq(from = 1, to = 5, by = 0.5)
my_seq2[1] 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0Here we’ve used the arguments from = and to = to define the limits of the sequence and the by = argument to specify the increment of the sequence. Play around with other values for these arguments to see their effect.
The rep() function allows you to replicate (repeat) values a specified number of times. To repeat the value 2, 10 times
my_seq3 <- rep(2, times = 10) # repeats 2, 10 times
my_seq3 [1] 2 2 2 2 2 2 2 2 2 2You can also repeat non-numeric values
my_seq4 <- rep("abc", times = 3) # repeats ‘abc’ 3 times
my_seq4[1] "abc" "abc" "abc"or each element of a series
my_seq5 <- rep(1:5, times = 3) # repeats the series 1 to
# 5, 3 times
my_seq5 [1] 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5or elements of a series
my_seq6 <- rep(1:5, each = 3) # repeats each element of the
# series 3 times
my_seq6 [1] 1 1 1 2 2 2 3 3 3 4 4 4 5 5 5We can also repeat a non-sequential series
[1] 3 3 3 1 1 1 10 10 10 7 7 7Note in the code above how we’ve used the c() function inside the rep() function. Nesting functions allows us to build quite complex commands within a single line of code and is a very common practice when using R. However, care needs to be taken as too many nested functions can make your code quite difficult for others to understand (or yourself some time in the future!). We could rewrite the code above to explicitly separate the two different steps to generate our vector. Either approach will give the same result, you just need to use your own judgement as to which is more readable.
Manipulating, summarising and sorting data using R is an important skill to master but one which many people find a little confusing at first. We’ll go through a few simple examples here using vectors to illustrate some important concepts but will build on this in much more detail in Chapter 3 where we will look at more complicated (and useful) data structures.
To extract (also known as indexing or subscripting) one or more values (more generally known as elements) from a vector we use the square bracket [ ] notation. The general approach is to name the object you wish to extract from, then a set of square brackets with an index of the element you wish to extract contained within the square brackets. This index can be a position or the result of a logical test.
To extract elements based on their position we simply write the position inside the [ ]. For example, to extract the 3rd value of my_vec
my_vec # remind ourselves what my_vec looks like[1] 2 3 1 6 4 3 3 7my_vec[3] # extract the 3rd value[1] 1# if you want to store this value in another object
val_3 <- my_vec[3]
val_3[1] 1Note that the positional index starts at 1 rather than 0 like some other other programming languages (i.e. Python).
We can also extract more than one value by using the c() function inside the square brackets. Here we extract the 1st, 5th, 6th and 8th element from the my_vec object
my_vec[c(1, 5, 6, 8)][1] 2 4 3 7Or we can extract a range of values using the : notation. To extract the values from the 3rd to the 8th elements
my_vec[3:8][1] 1 6 4 3 3 7Another really useful way to extract data from a vector is to use a logical expression as an index. For example, to extract all elements with a value greater than 4 in the vector my_vec
my_vec[my_vec > 4][1] 6 7Here, the logical expression is my_vec > 4 and R will only extract those elements that satisfy this logical condition. So how does this actually work? If we look at the output of just the logical expression without the square brackets you can see that R returns a vector containing either TRUE or FALSE which correspond to whether the logical condition is satisfied for each element. In this case only the 4th and 8th elements return a TRUE as their value is greater than 4.
my_vec > 4[1] FALSE FALSE FALSE TRUE FALSE FALSE FALSE TRUESo what R is actually doing under the hood is equivalent to
my_vec[c(FALSE, FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, TRUE)][1] 6 7and only those element that are TRUE will be extracted.
In addition to the < and > operators you can also use composite operators to increase the complexity of your expressions. For example the expression for ‘greater or equal to’ is >=. To test whether a value is equal to a value we need to use a double equals symbol == and for ‘not equal to’ we use != (the ! symbol means ‘not’).
my_vec[my_vec >= 4] # values greater or equal to 4[1] 6 4 7my_vec[my_vec < 4] # values less than 4[1] 2 3 1 3 3my_vec[my_vec <= 4] # values less than or equal to 4[1] 2 3 1 4 3 3my_vec[my_vec == 4] # values equal to 4[1] 4my_vec[my_vec != 4] # values not equal to 4[1] 2 3 1 6 3 3 7We can also combine multiple logical expressions using Boolean expressions. In R the & symbol means AND and the | symbol means OR. For example, to extract values in my_vec which are less than 6 AND greater than 2
val26 <- my_vec[my_vec < 6 & my_vec > 2]
val26[1] 3 4 3 3or extract values in my_vec that are greater than 6 OR less than 3
val63 <- my_vec[my_vec > 6 | my_vec < 3]
val63[1] 2 1 7We can change the values of some elements in a vector using our [ ] notation in combination with the assignment operator <-. For example, to replace the 4th value of our my_vec object from 6 to 500
my_vec[4] <- 500
my_vec[1] 2 3 1 500 4 3 3 7We can also replace more than one value or even replace values based on a logical expression
# replace the 6th and 7th element with 100
my_vec[c(6, 7)] <- 100
my_vec[1] 2 3 1 500 4 100 100 7# replace element that are less than or equal to 4 with 1000
my_vec[my_vec <= 4] <- 1000
my_vec[1] 1000 1000 1000 500 1000 100 100 7In addition to extracting particular elements from a vector we can also order the values contained in a vector. To sort the values from lowest to highest value we can use the sort() function
vec_sort <- sort(my_vec)
vec_sort[1] 7 100 100 500 1000 1000 1000 1000To reverse the sort, from highest to lowest, we can either include the decreasing = TRUE argument when using the sort() function
vec_sort2 <- sort(my_vec, decreasing = TRUE)
vec_sort2[1] 1000 1000 1000 1000 500 100 100 7or first sort the vector using the sort() function and then reverse the sorted vector using the rev() function. This is another example of nesting one function inside another function.
Whilst sorting a single vector is fun, perhaps a more useful task would be to sort one vector according to the values of another vector. To do this we should use the order() function in combination with [ ]. To demonstrate this let’s create a vector called height containing the height of 5 different people and another vector called p.names containing the names of these people (so Joanna is 180 cm, Charlotte is 155 cm etc)
height <- c(180, 155, 160, 167, 181)
height[1] 180 155 160 167 181p.names <- c("Joanna", "Charlotte", "Helen", "Karen", "Amy")
p.names[1] "Joanna" "Charlotte" "Helen" "Karen" "Amy" Our goal is to order the people in p.names in ascending order of their height. The first thing we’ll do is use the order() function with the height variable to create a vector called height_ord
height_ord <- order(height)
height_ord[1] 2 3 4 1 5OK, what’s going on here? The first value, 2, (remember ignore [1]) should be read as ‘the smallest value of height is the second element of the height vector’. If we check this by looking at the height vector above, you can see that element 2 has a value of 155, which is the smallest value. The second smallest value in height is the 3rd element of height, which when we check is 160 and so on. The largest value of height is element 5 which is 181. Now that we have a vector of the positional indices of heights in ascending order (height_ord), we can extract these values from our p.names vector in this order
names_ord <- p.names[height_ord]
names_ord[1] "Charlotte" "Helen" "Karen" "Joanna" "Amy" You’re probably thinking ‘what’s the use of this?’ Well, imagine you have a dataset which contains two columns of data and you want to sort each column. If you just use sort() to sort each column separately, the values of each column will become uncoupled from each other. By using the ‘order()’ on one column, a vector of positional indices is created of the values of the column in ascending order This vector can be used on the second column, as the index of elements which will return a vector of values based on the first column. In all honestly, when you have multiple related vectors you need to use a data.frame type of object (see Chapter 3) instead of multiple independent vectors.
One of the great things about R functions is that most of them are vectorised. This means that the function will operate on all elements of a vector without needing to apply the function on each element separately. For example, to multiple each element of a vector by 5 we can simply use
# create a vector
my_vec2 <- c(3, 5, 7, 1, 9, 20)
# multiply each element by 5
my_vec2 * 5[1] 15 25 35 5 45 100Or we can add the elements of two or more vectors
# create a second vector
my_vec3 <- c(17, 15, 13, 19, 11, 0)
# add both vectors
my_vec2 + my_vec3[1] 20 20 20 20 20 20# multiply both vectors
my_vec2 * my_vec3[1] 51 75 91 19 99 0However, you must be careful when using vectorisation with vectors of different lengths as R will quietly recycle the elements in the shorter vector rather than throw a wobbly (error).
# create a third vector
my_vec4 <- c(1, 2)
# add both vectors - quiet recycling!
my_vec2 + my_vec4[1] 4 7 8 3 10 22In R, missing data is usually represented by an NA symbol meaning ‘Not Available’. Data may be missing for a whole bunch of reasons, maybe your machine broke down, maybe you broke down, maybe the weather was too bad to collect data on a particular day etc etc. Missing data can be a pain in the proverbial both from an R perspective and also a statistical perspective. From an R perspective missing data can be problematic as different functions deal with missing data in different ways. For example, let’s say we collected air temperature readings over 10 days, but our thermometer broke on day 2 and again on day 9 so we have no data for those days
temp <- c(7.2, NA, 7.1, 6.9, 6.5, 5.8, 5.8, 5.5, NA, 5.5)
temp [1] 7.2 NA 7.1 6.9 6.5 5.8 5.8 5.5 NA 5.5We now want to calculate the mean temperature over these days using the mean() function
mean_temp <- mean(temp)
mean_temp[1] NAIf a vector has a missing value then the only possible value to return when calculating a mean is NA. R doesn’t know that you perhaps want to ignore the NA values (R can’t read your mind - yet!). If we look at the help file (using ?mean - see the next section Section 2.6 for more details) associated with the mean() function we can see there is an argument na.rm = which is set to FALSE by default.
na.rm - a logical value indicating whether NA values should be stripped before the computation proceeds.
If we change this argument to na.rm = TRUE when we use the mean() function this will allow us to ignore the NA values when calculating the mean
mean_temp <- mean(temp, na.rm = TRUE)
mean_temp[1] 6.2875It’s important to note that the NA values have not been removed from our temp object (that would be bad practice), rather the mean() function has just ignored them. The point of the above is to highlight how we can change the default behaviour of a function using an appropriate argument. The problem is that not all functions will have an na.rm = argument, they might deal with NA values differently. However, the good news is that every help file associated with any function will always tell you how missing data are handled by default.
This book is intended as a relatively brief introduction to R and as such you will soon be using functions and packages that go beyond this scope of this introductory text. Fortunately, one of the strengths of R is its comprehensive and easily accessible help system and wealth of online resources where you can obtain further information.
To access R’s built-in help facility to get information on any function simply use the help() function. For example, to open the help page for our friend the mean() function.
help("mean")or you can use the equivalent shortcut
?meanthe help page is displayed in the ‘Help’ tab in the Files pane (usually in the bottom right of RStudio)
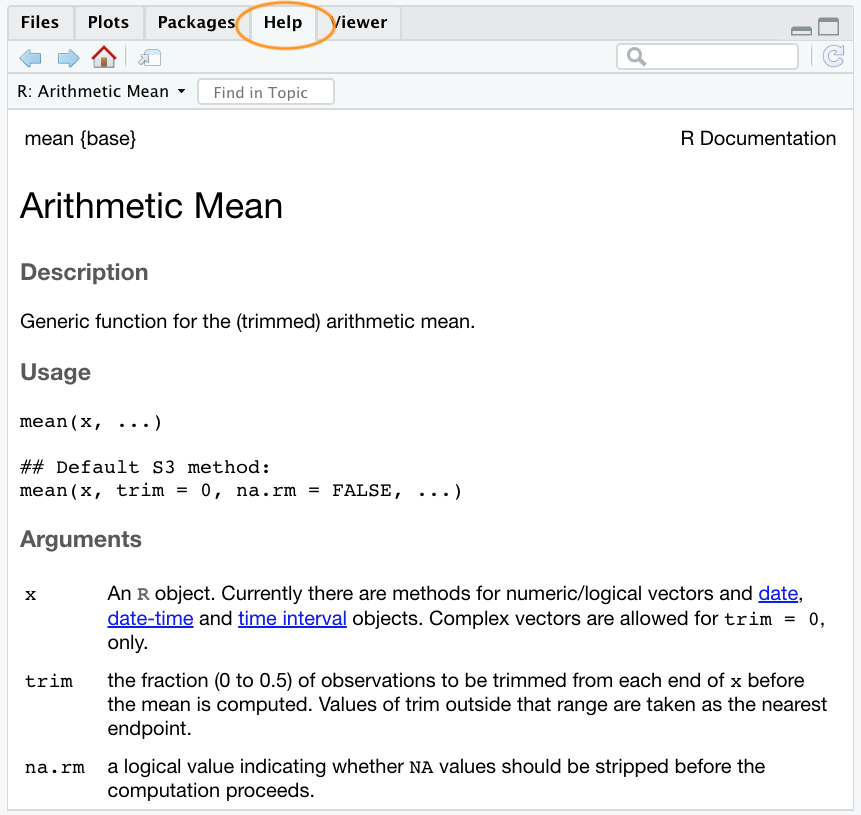
mean() function in RStudio Help pane
Admittedly the help files can seem anything but helpful when you first start using R. This is probably because they’re written in a very concise manner and the language used is often quite technical and full of jargon. Having said that, you do get used to this and will over time even come to appreciate a certain beauty in their brevity (honest!). One of the great things about the help files is that they all have a very similar structure regardless of the function. This makes it easy to navigate through the file to find exactly what you need.
The first line of the help document contains information such as the name of the function and the package where the function can be found. There are also other headings that provide more specific information such as
| Headings | Description |
|---|---|
| Description: | gives a brief description of the function and what it does. |
| Usage: | gives the name of the arguments associated with the function and possible default values. |
| Arguments: | provides more detail regarding each argument and what they do. |
| Details: | gives further details of the function if required. |
| Value: | if applicable, gives the type and structure of the object returned by the function or the operator. |
| See Also: | provides information on other help pages with similar or related content. |
| Examples: | gives some examples of using the function. |
The Examples are are really helpful, all you need to do is copy and paste them into the console to see what happens. You can also access examples at any time by using the example() function (i.e. example("mean"))
The help() function is useful if you know the name of the function. If you’re not sure of the name, but can remember a key word then you can search R’s help system using the help.search() function.
help.search("mean")or you can use the equivalent shortcut
??meanThe results of the search will be displayed in RStudio under the ‘Help’ tab as before. The help.search() function searches through the help documentation, code demonstrations and package vignettes and displays the results as clickable links for further exploration.
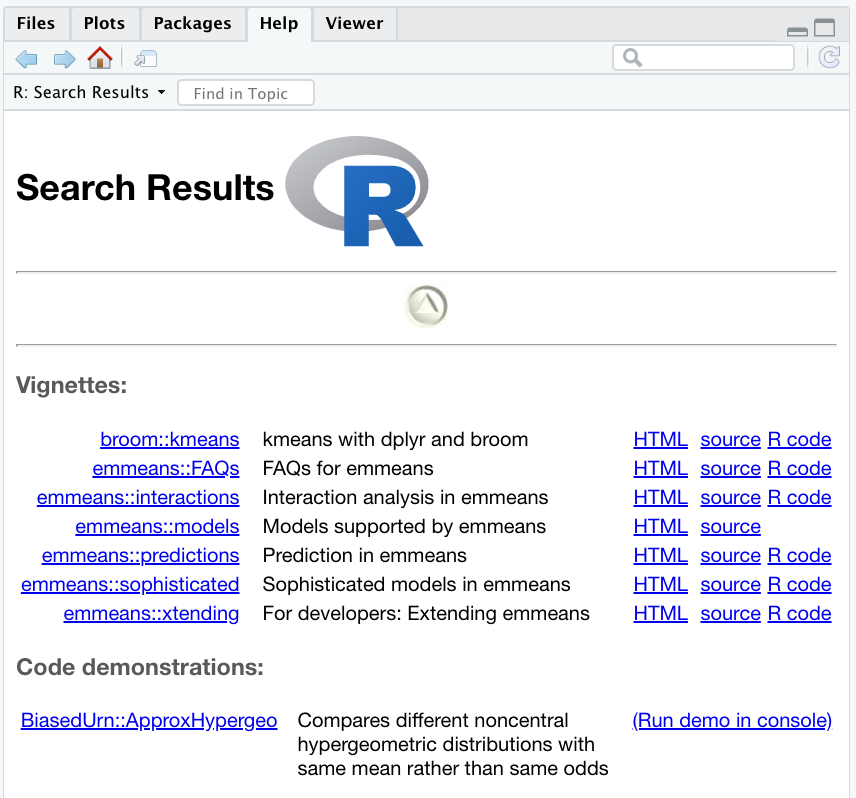
help.search() function in RStudio
Another useful function is apropos(). This function can be used to list all functions containing a specified character string. For example, to find all functions with mean in their name
apropos("mean") [1] ".colMeans" ".rowMeans" "colMeans" "kmeans"
[5] "mean" "mean_temp" "mean.Date" "mean.default"
[9] "mean.difftime" "mean.POSIXct" "mean.POSIXlt" "rowMeans"
[13] "vec_mean" "weighted.mean"You can then bring up the help file for the relevant function.
help("kmeans")Another function is RSiteSearch() which enables you to search for keywords and phrases in function help pages and vignettes for all CRAN packages. This function allows you to access the search engine of the R website https://www.r-project.org/search.html directly from the Console with the results displayed in your web browser.
RSiteSearch("regression")There really has never been a better time to start learning R. There are a plethora of freely available online resources ranging from whole courses to subject specific tutorials and mailing lists. There are also plenty of paid for options if that’s your thing but unless you’ve money to burn there really is no need to part with your hard earned cash. Some resources we have found helpful are listed below.
[Internet search]: Use your favourite search engine (google, ecosia, duckduckgo, … )for any error messages you get. It’s not cheating and everyone does it! You’ll be surprised how many other people have probably had the same problem and solved it.
Stack Overflow: There are many thousands of questions relevant to R on Stack Overflow. Here are the most popular ones, ranked by vote. Make sure you search for similar questions before asking your own, and make sure you include a reproducible example to get the most useful advice. A reproducible example is a minimal example that lets others who are trying to help you to see the error themselves.
Your approach to saving work in R and RStudio depends on what you want to save. Most of the time the only thing you will need to save is the R code in your script(s). Remember your script is a reproducible record of everything you’ve done so all you need to do is open up your script in a new RStudio session and source it into the R Console and you’re back to where you left off.
Unless you’ve followed our suggestion about changing the default settings for RStudio Projects (see Section 1.5) you will be asked whether you want to save your workspace image every time you exit RStudio. We suggest that 99.9% of the time that you don’t want do this. By starting with a clean RStudio session each time we come back to our analysis we can be sure to avoid any potential conflicts with things we’ve done in previous sessions.
There are, however, some occasions when saving objects you’ve created in R is useful. For example, let’s say you’re creating an object that takes hours (even days) of computational time to generate. It would be extremely inconvenient to have to wait all this time each time you come back to your analysis (although we would suggest exporting this to an external file is a better solution). In this case we can save this object as an external .RData file which we can load back into RStudio the next time we want to use it. To save an object to an .RData file you can use the save() function (notice we don’t need to use the assignment operator here)
save(nameOfObject, file = "name_of_file.RData")or if you want to save all of the objects in your workspace into a single .RData file use the save.image() function
save.image(file = "name_of_file.RData")To load your .RData file back into RStudio use the load() function
load(file = "name_of_file.RData")The base installation of R comes with many useful packages as standard. These packages will contain many of the functions you will use on a daily basis. However, as you start using R for more diverse projects (and as your own use of R evolves) you will find that there comes a time when you will need to extend R’s capabilities. Happily, many thousands of R users have developed useful code and shared this code as installable packages. You can think of a package as a collection of functions, data and help files collated into a well defined standard structure which you can download and install in R. These packages can be downloaded from a variety of sources but the most popular are CRAN, Bioconductor and GitHub. Currently, CRAN hosts over 15000 packages and is the official repository for user contributed R packages. Bioconductor provides open source software oriented towards bioinformatics and hosts over 1800 R packages. GitHub is a website that hosts git repositories for all sorts of software and projects (not just R). Often, cutting edge development versions of R packages are hosted on GitHub so if you need all the new bells and whistles then this may be an option. However, a potential downside of using the development version of an R package is that it might not be as stable as the version hosted on CRAN (it’s in development!) and updating packages won’t be automatic.
Once you have installed a package onto your computer it is not immediately available for you to use. To use a package you first need to load the package by using the library() function. For example, to load the remotes 📦 package you previously installed
The library() function will also load any additional packages required and may print out additional package information. It is important to realize that every time you start a new R session (or restore a previously saved session) you need to load the packages you will be using. We tend to put all our library() statements required for our analysis near the top of our R scripts to make them easily accessible and easy to add to as our code develops. If you try to use a function without first loading the relevant R package you will receive an error message that R could not find the function. For example, if you try to use the install_github() function without loading the remotes 📦 package first you will receive the following error
install_github("tidyverse/dplyr")
# Error in install_github("tidyverse/dplyr") :
# could not find function "install_github"Sometimes it can be useful to use a function without first using the library() function. If, for example, you will only be using one or two functions in your script and don’t want to load all of the other functions in a package then you can access the function directly by specifying the package name followed by two colons and then the function name
remotes::install_github("tidyverse/dplyr")This is how we were able to use the install() and install_github() functions above without first loading the packages BiocManager 📦 and remotes 📦 . Most of the time we recommend using the library() function.
To install a package from CRAN you can use the install.packages() function. For example if you want to install the remotes package enter the following code into the Console window of RStudio (note: you will need a working internet connection to do this)
install.packages("remotes", dependencies = TRUE)You may be asked to select a CRAN mirror, just select ‘0-cloud’ or a mirror near to your location. The dependencies = TRUE argument ensures that additional packages that are required will also be installed.
It’s good practice to regularly update your previously installed packages to get access to new functionality and bug fixes. To update CRAN packages you can use the update.packages() function (you will need a working internet connection for this)
update.packages(ask = FALSE)The ask = FALSE argument avoids having to confirm every package download which can be a pain if you have many packages installed.
To install packages from Bioconductor the process is a little different. You first need to install the BiocManager 📦 package. You only need to do this once unless you subsequently reinstall or upgrade R
install.packages("BiocManager", dependencies = TRUE)Once the BiocManager📦 package has been installed you can either install all of the ‘core’ Bioconductor packages with
BiocManager::install()or install specific packages such as the GenomicRanges 📦 and edgeR 📦 packages
To update Bioconductor packages just use the BiocManager::install() function again
BiocManager::install(ask = FALSE)Again, you can use the ask = FALSE argument to avoid having to confirm every package download.
There are multiple options for installing packages hosted on GitHub. Perhaps the most efficient method is to use the install_github() function from the remotes 📦 package (you installed this package previously (Section 2.8.2.1)). Before you use the function you will need to know the GitHub username of the repository owner and also the name of the repository. For example, the development version of dplyr 📦 from Hadley Wickham is hosted on the tidyverse GitHub account and has the repository name ‘dplyr’ (just search for ‘github dplyr’). To install this version from GitHub use
remotes::install_github("tidyverse/dplyr")The safest way (that we know of) to update a package installed from GitHub is to just reinstall it using the above command.