15 Generalized linear model, glm
15.1 Lecture
m1 <- glm(fish ~ french_captain, data = dads_joke, family = poisson)15.1.1 Distributions
15.1.1.1 Continuous linear
- Gaussian
15.1.1.2 Count data
- poisson
- negative binomial
- quasi-poisson
- generalized poisson
- conway-maxwell poisson
15.1.1.3 censored distribution
15.1.1.4 zero-inflated / hurdle distribution
- zero-inflated/zero-truncated poisson
- censored poisson
15.1.1.5 zero-truncated distribution
15.1.1.6 zero-one-inflated distribution
see https://cran.r-project.org/web/packages/brms/vignettes/brms_families.html see alo MCMCglmm coursenotes
for help on description and to add some plots about those distribution
15.2 Practical
Warning
This section need to be severely updated
15.2.1 Logistic regression
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsThis is DHARMa 0.4.6. For overview type '?DHARMa'. For recent changes, type news(package = 'DHARMa')
bubble <- data.frame(age = rep(2:16, 2),
reproduction = rep(0:1, each = 15),
size = c(table(mouflonc$age, mouflonc$reproduction)))
bubble$size <- ifelse(bubble$size == 0 , NA, bubble$size)
ggplot(data = bubble, aes(x = age, y = reproduction))+
geom_point(aes(size = size*10))Warning: Removed 7 rows containing missing values or values outside the scale range
(`geom_point()`).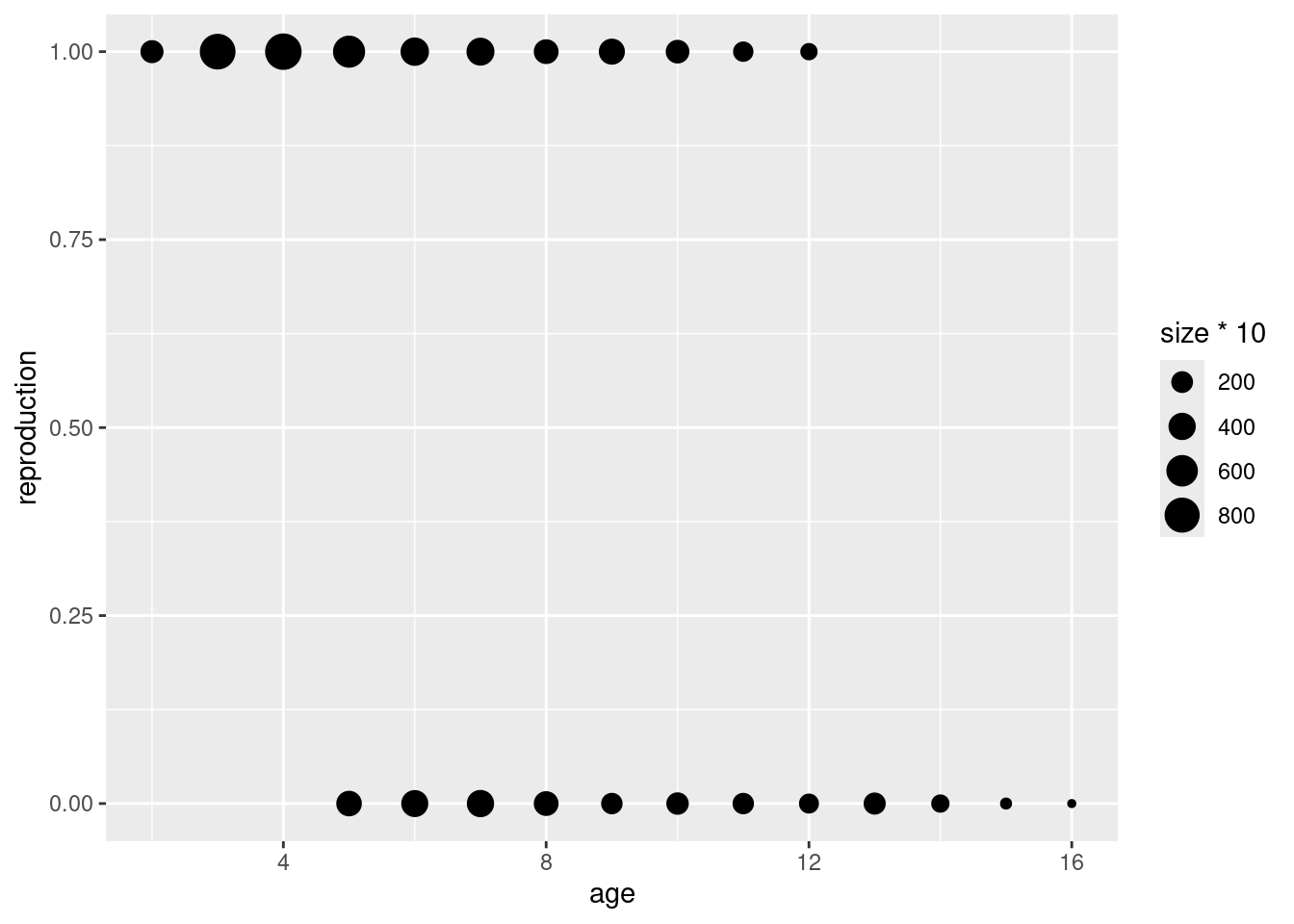
Call:
glm(formula = reproduction ~ age, family = binomial, data = mouflonc)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 3.19921 0.25417 12.59 <2e-16 ***
age -0.36685 0.03287 -11.16 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 928.86 on 715 degrees of freedom
Residual deviance: 767.51 on 714 degrees of freedom
(4 observations deleted due to missingness)
AIC: 771.51
Number of Fisher Scoring iterations: 4simulationOutput <- simulateResiduals(m1)
plot(simulationOutput)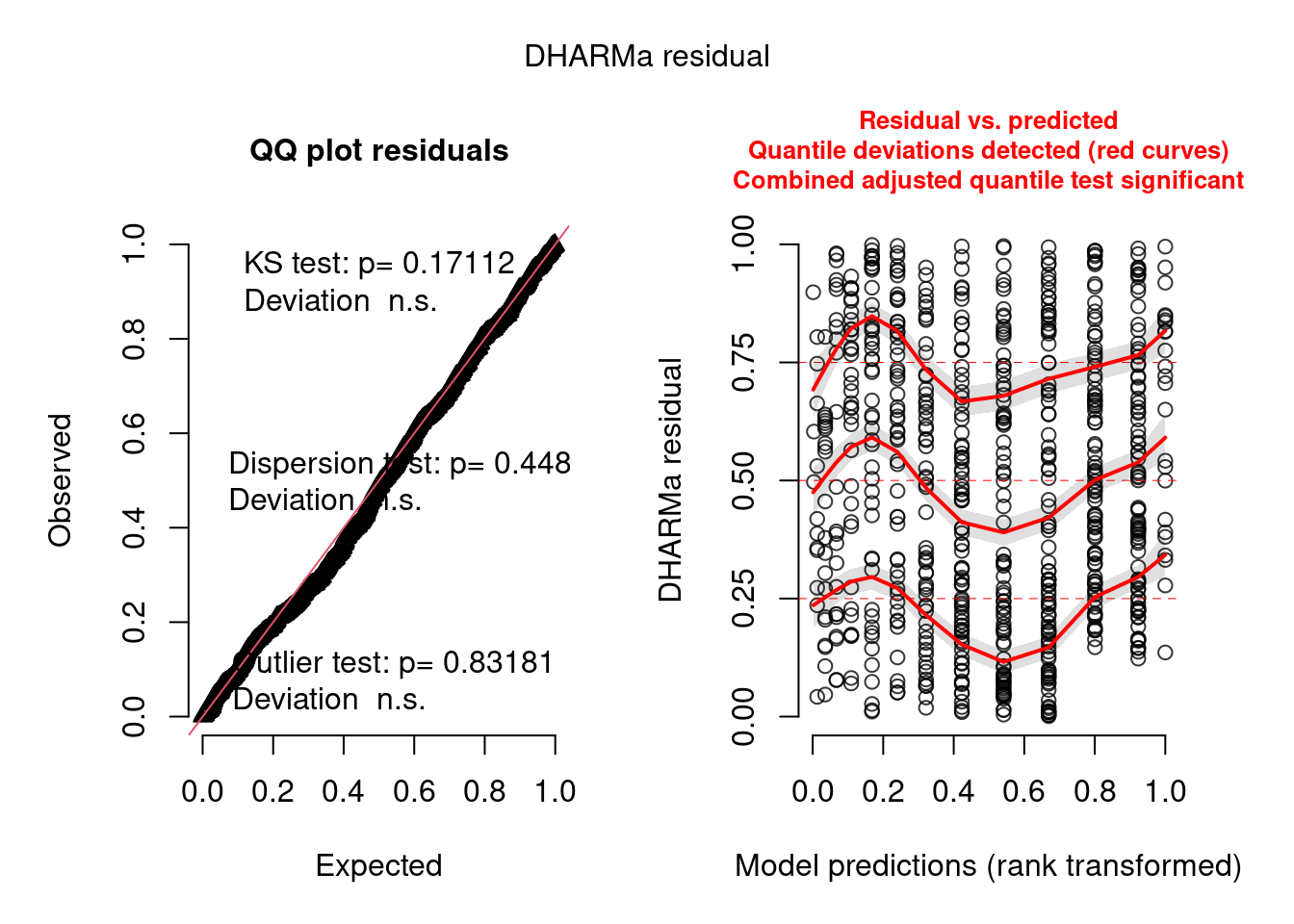
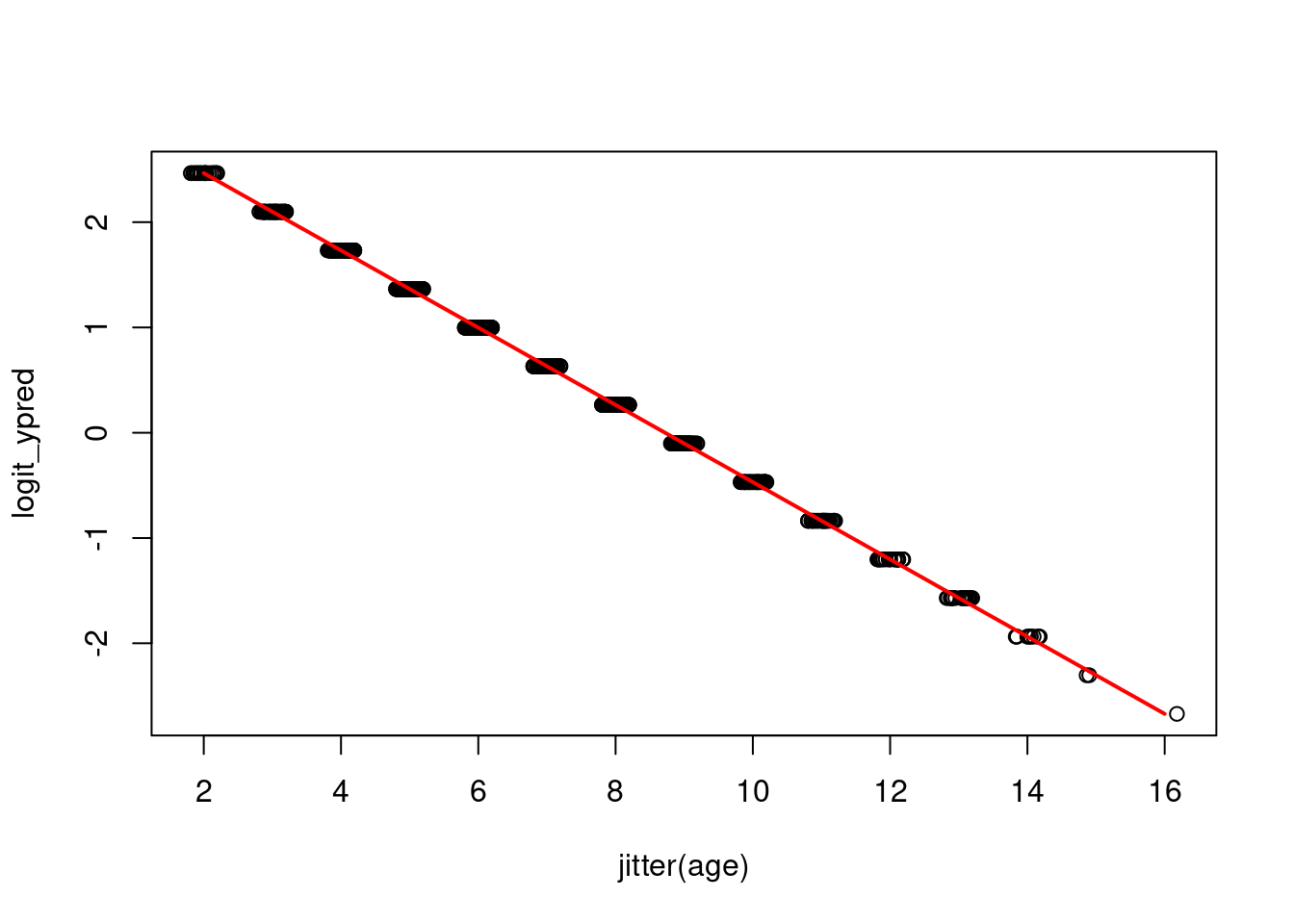
plotting the model prediction on the link (latent) scale
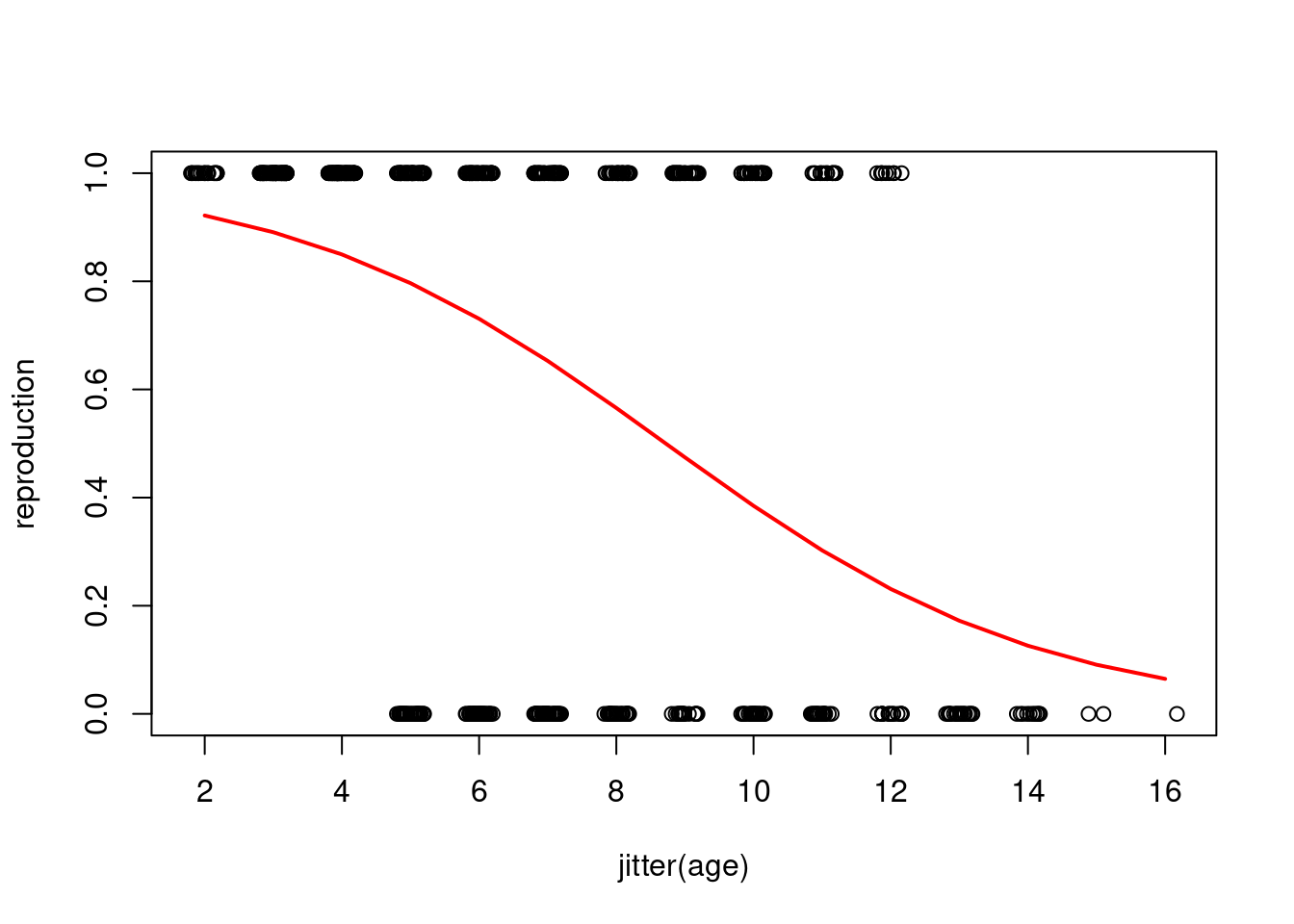
plotting on the observed scale
Enfin, pour se simplifier la vie, il est aussi possible de récupérer les valeurs prédites de y directement
Call:
glm(formula = reproduction ~ age + mass_sept + as.factor(sex_lamb) +
mass_gain + density + temp, family = binomial, data = mouflon)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.622007 1.943242 0.835 0.403892
age -0.148567 0.033597 -4.422 9.78e-06 ***
mass_sept 0.029878 0.016815 1.777 0.075590 .
as.factor(sex_lamb)1 -0.428169 0.166156 -2.577 0.009969 **
mass_gain -0.094828 0.026516 -3.576 0.000348 ***
density -0.018132 0.003518 -5.154 2.55e-07 ***
temp 0.037244 0.138712 0.269 0.788313
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 916.06 on 674 degrees of freedom
Residual deviance: 845.82 on 668 degrees of freedom
(45 observations deleted due to missingness)
AIC: 859.82
Number of Fisher Scoring iterations: 4check_model(m2)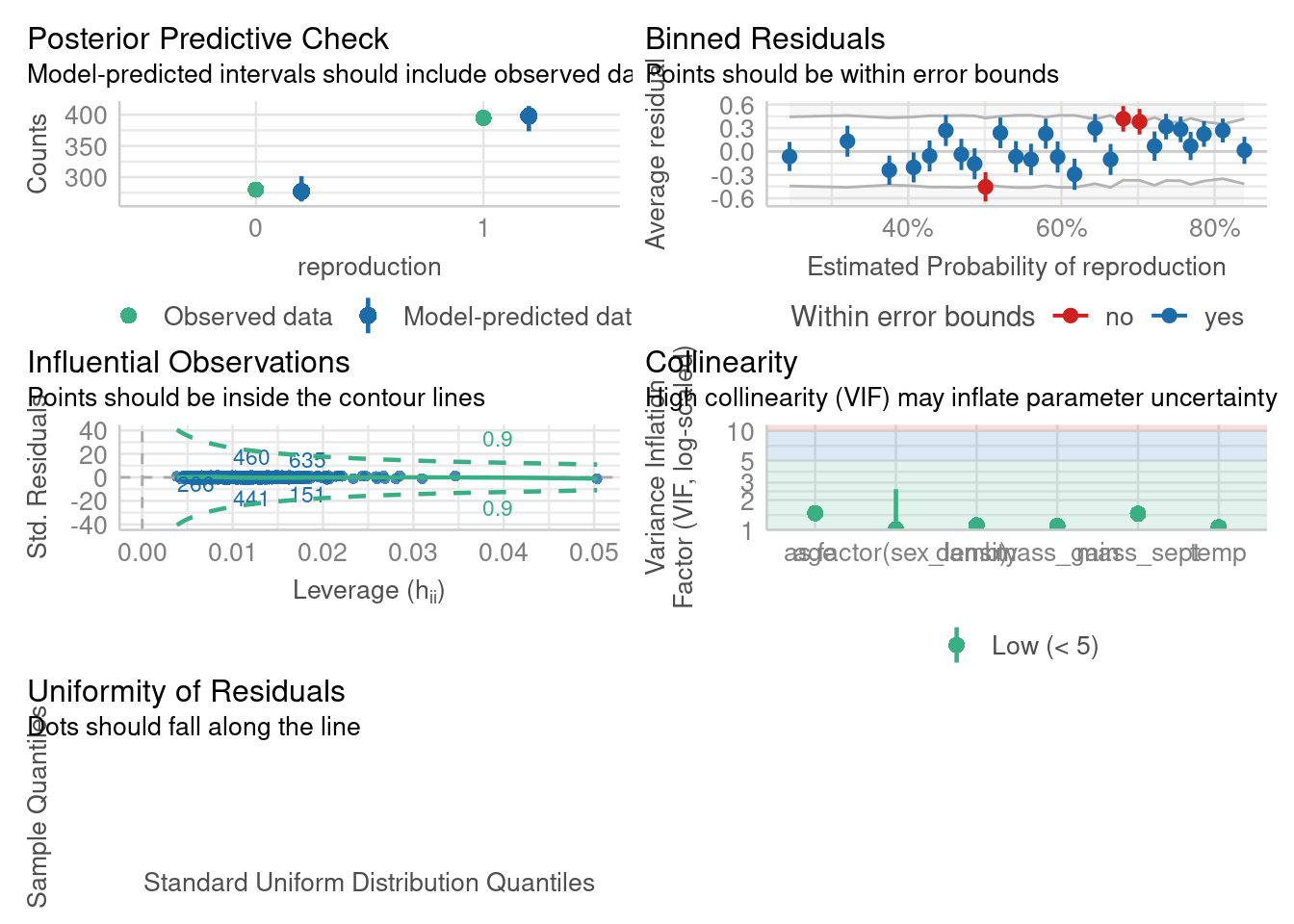
simulationOutput <- simulateResiduals(m2)
plot(simulationOutput)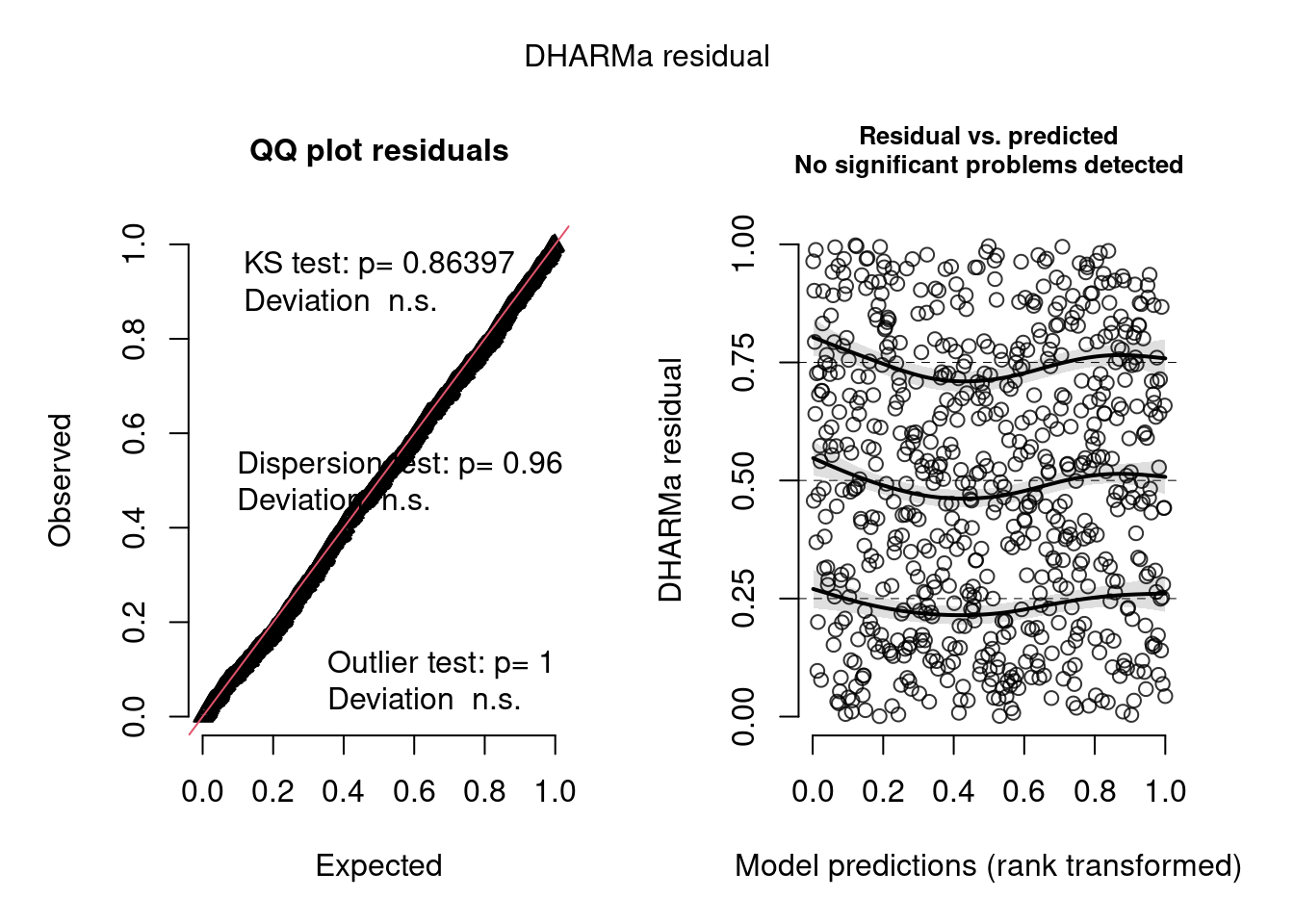
15.2.1.1 previous offspring sex effect
15.2.2 Poisson regression
data on galapagos islands species richness model of total number of species model of proportion of native model of density of species
Fit 3 models - model of total number of species - model of proportion of endemics to total - model of species density
#
gala <- read.delim2("data/gala.txt")
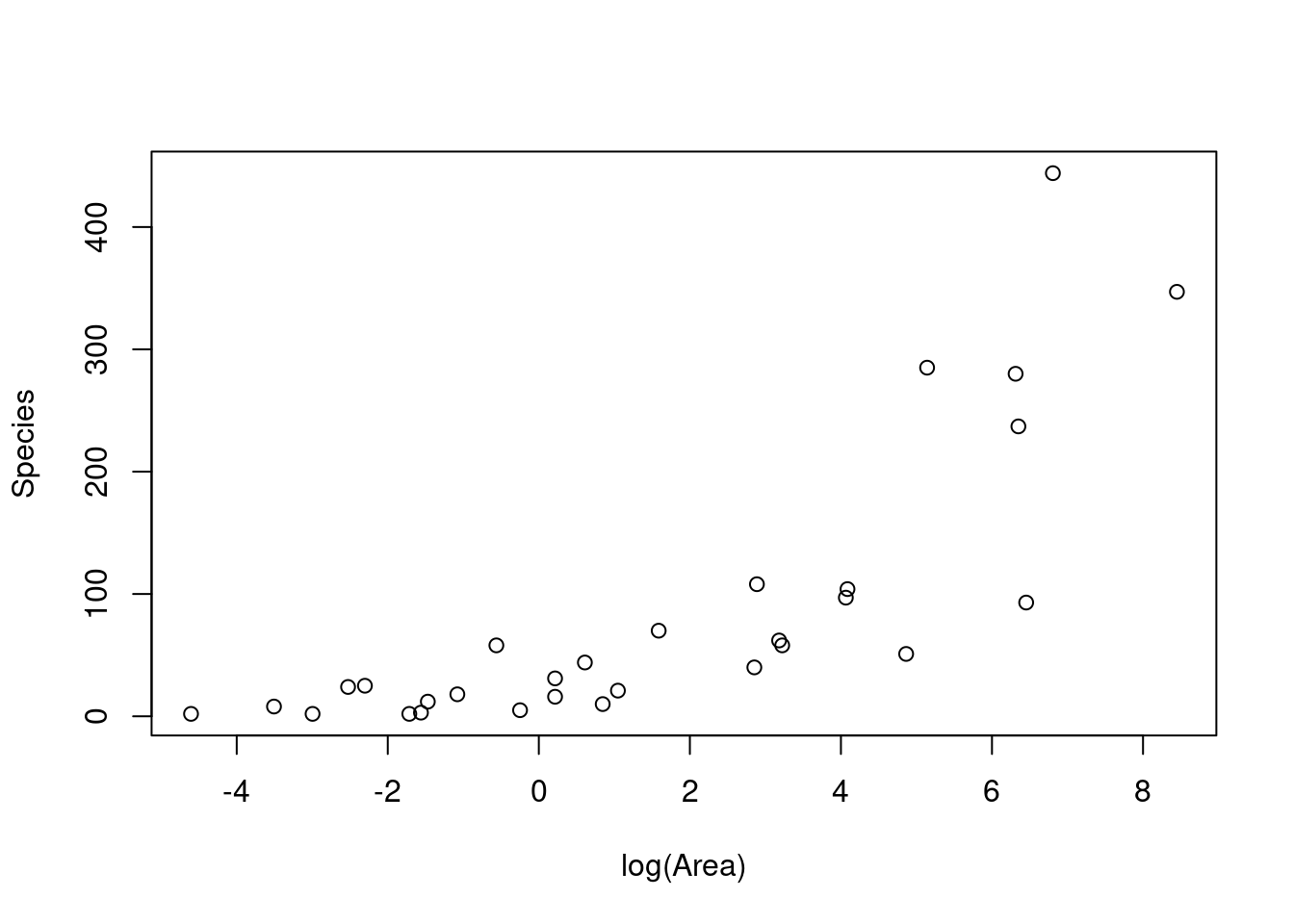
plot(Species ~ Area, gala)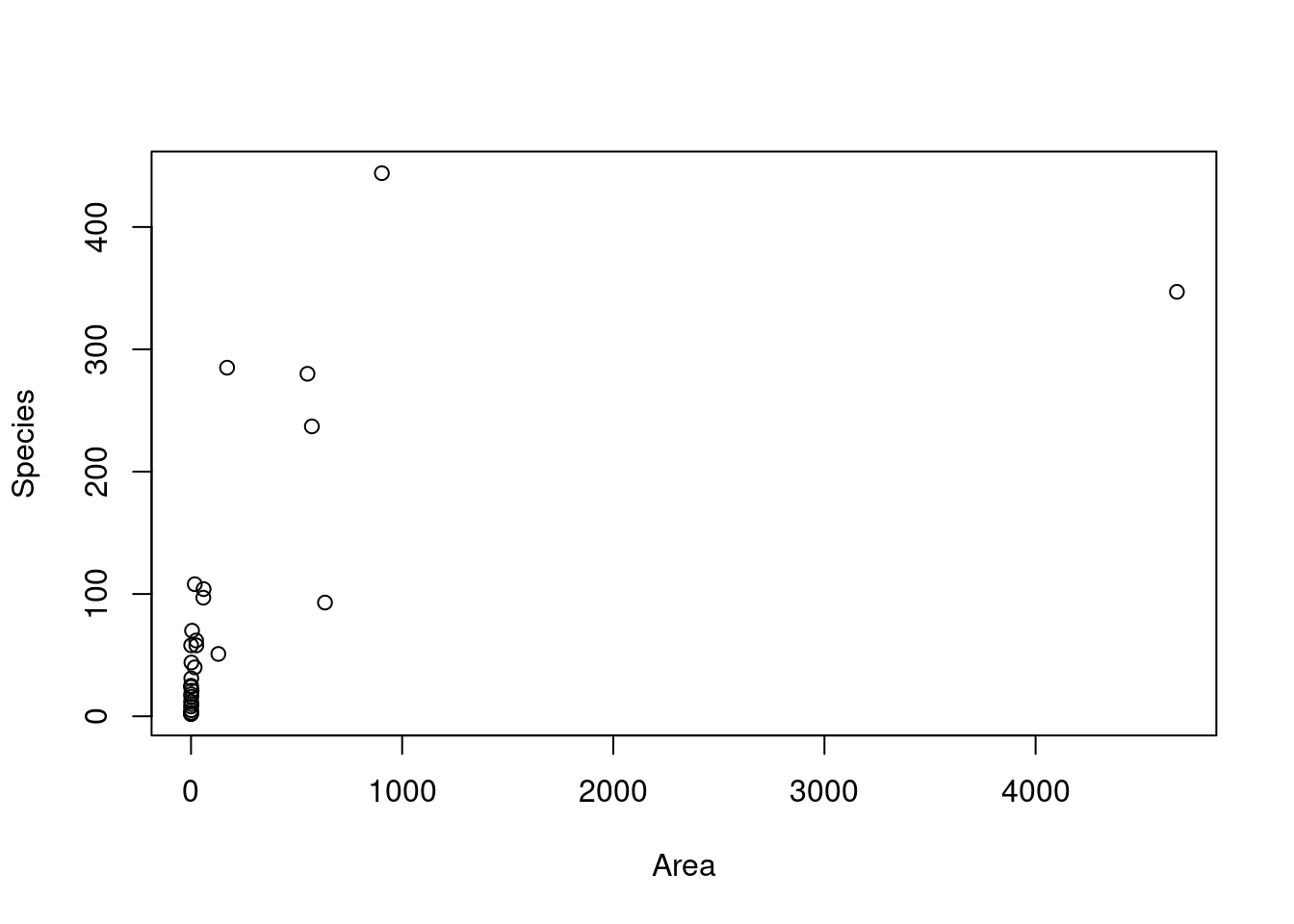
hist(gala$Species)
modpl <- glm(Species ~ Area + Elevation + Nearest, family=poisson, gala)
res <- simulateResiduals(modpl)
testDispersion(res)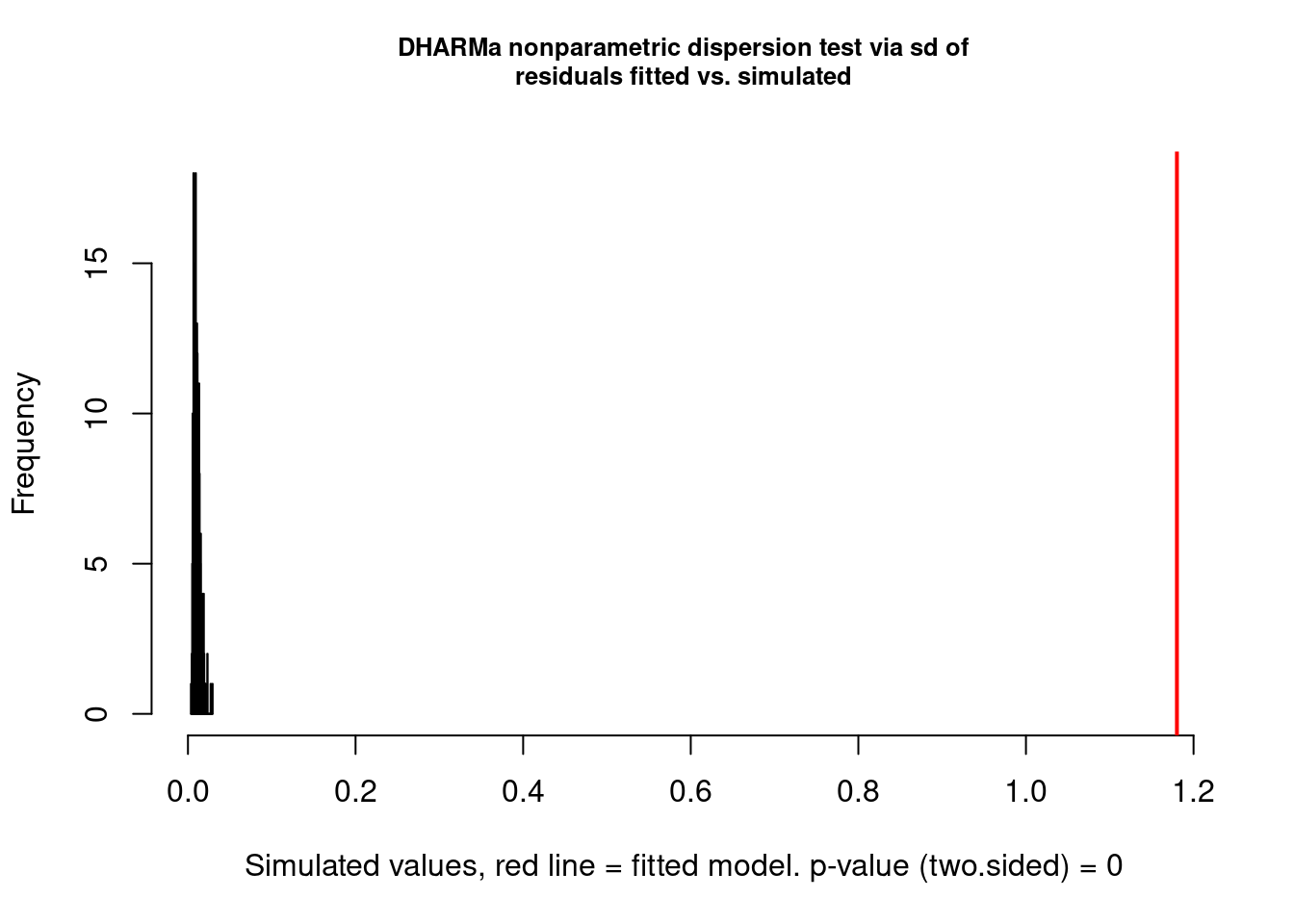
DHARMa nonparametric dispersion test via sd of residuals fitted vs.
simulated
data: simulationOutput
dispersion = 110.32, p-value < 2.2e-16
alternative hypothesis: two.sidedtestZeroInflation(res)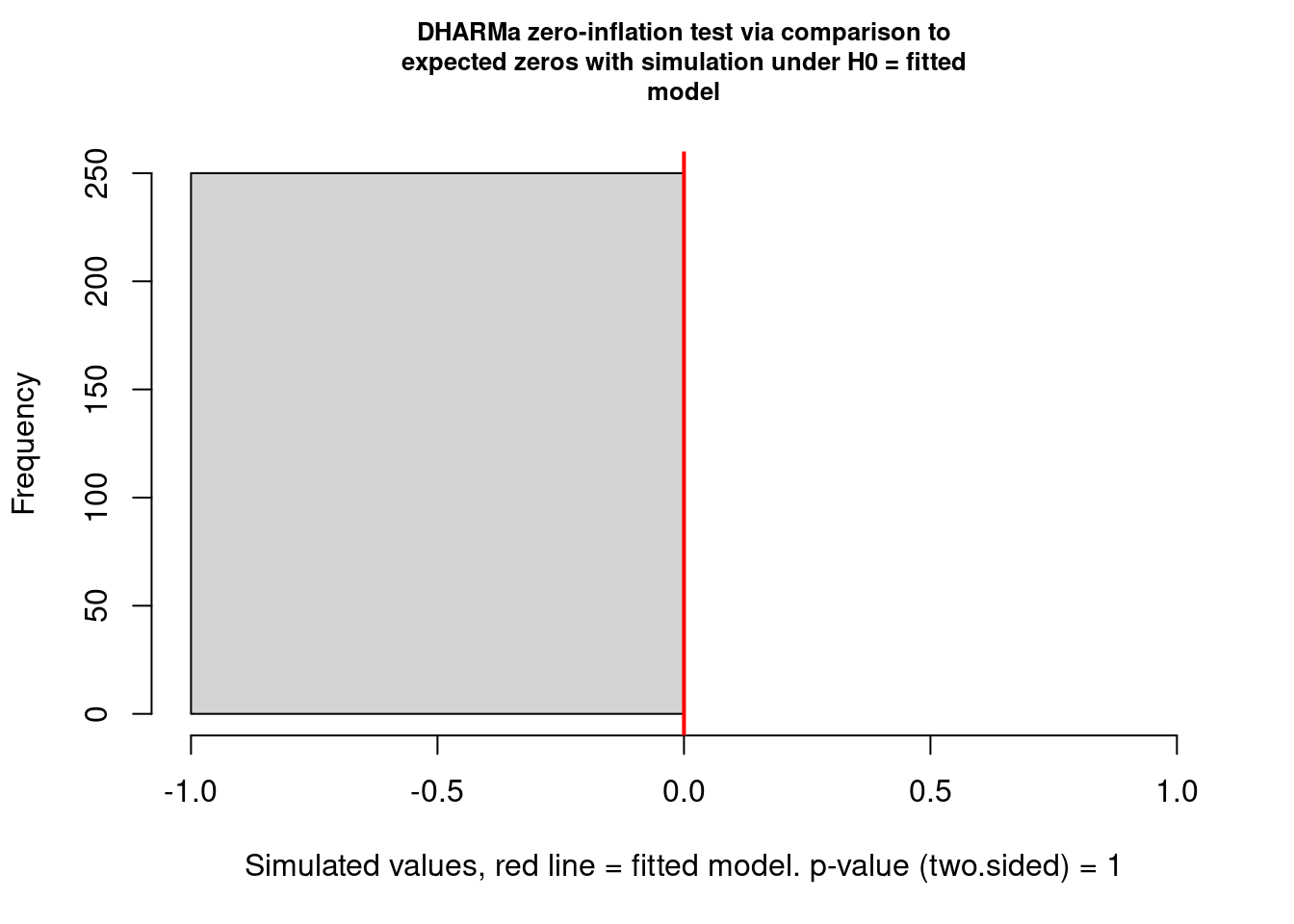
DHARMa zero-inflation test via comparison to expected zeros with
simulation under H0 = fitted model
data: simulationOutput
ratioObsSim = NaN, p-value = 1
alternative hypothesis: two.sided mean(gala$Species)[1] 85.23333 var(gala$Species)[1] 13140.74
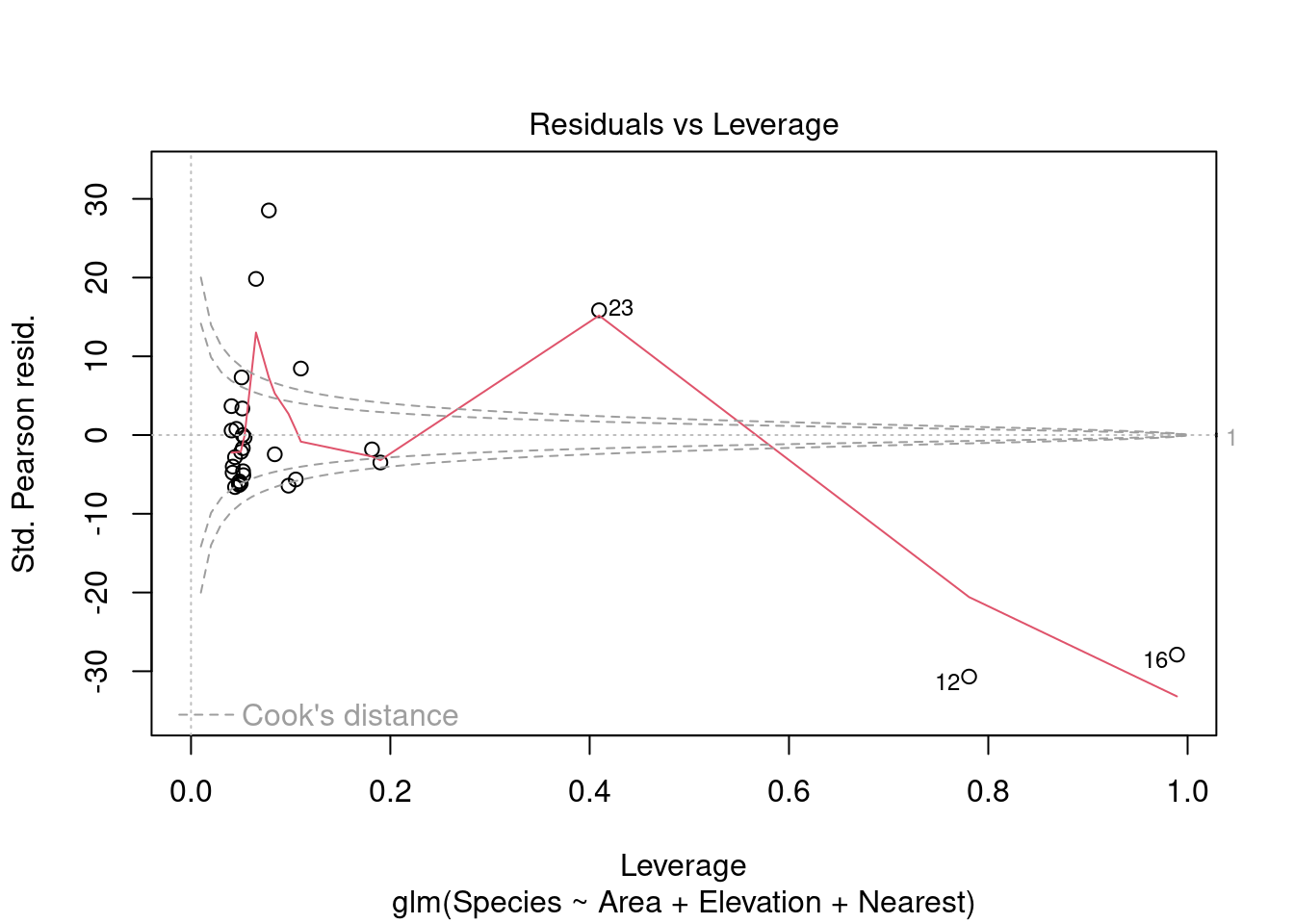
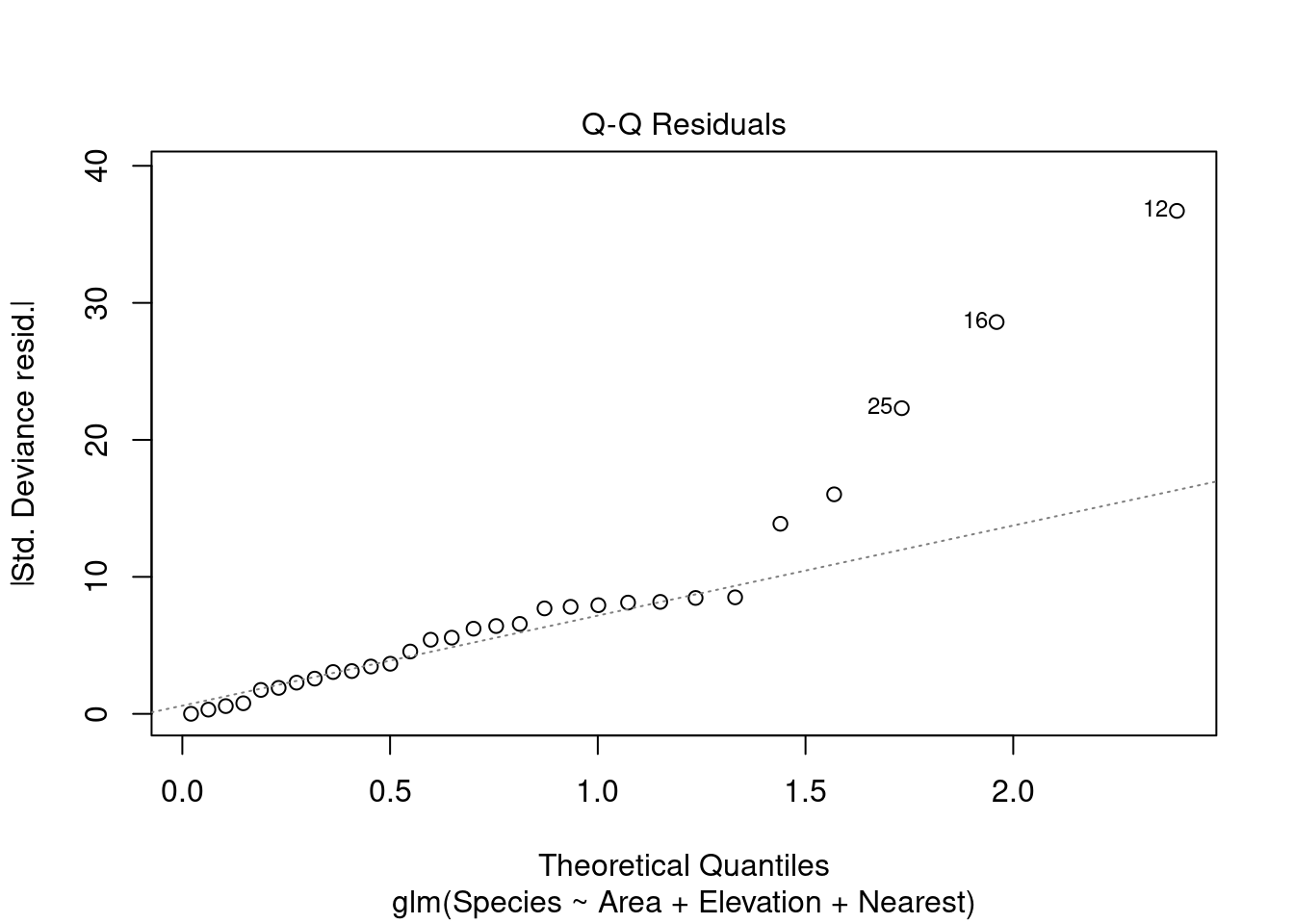
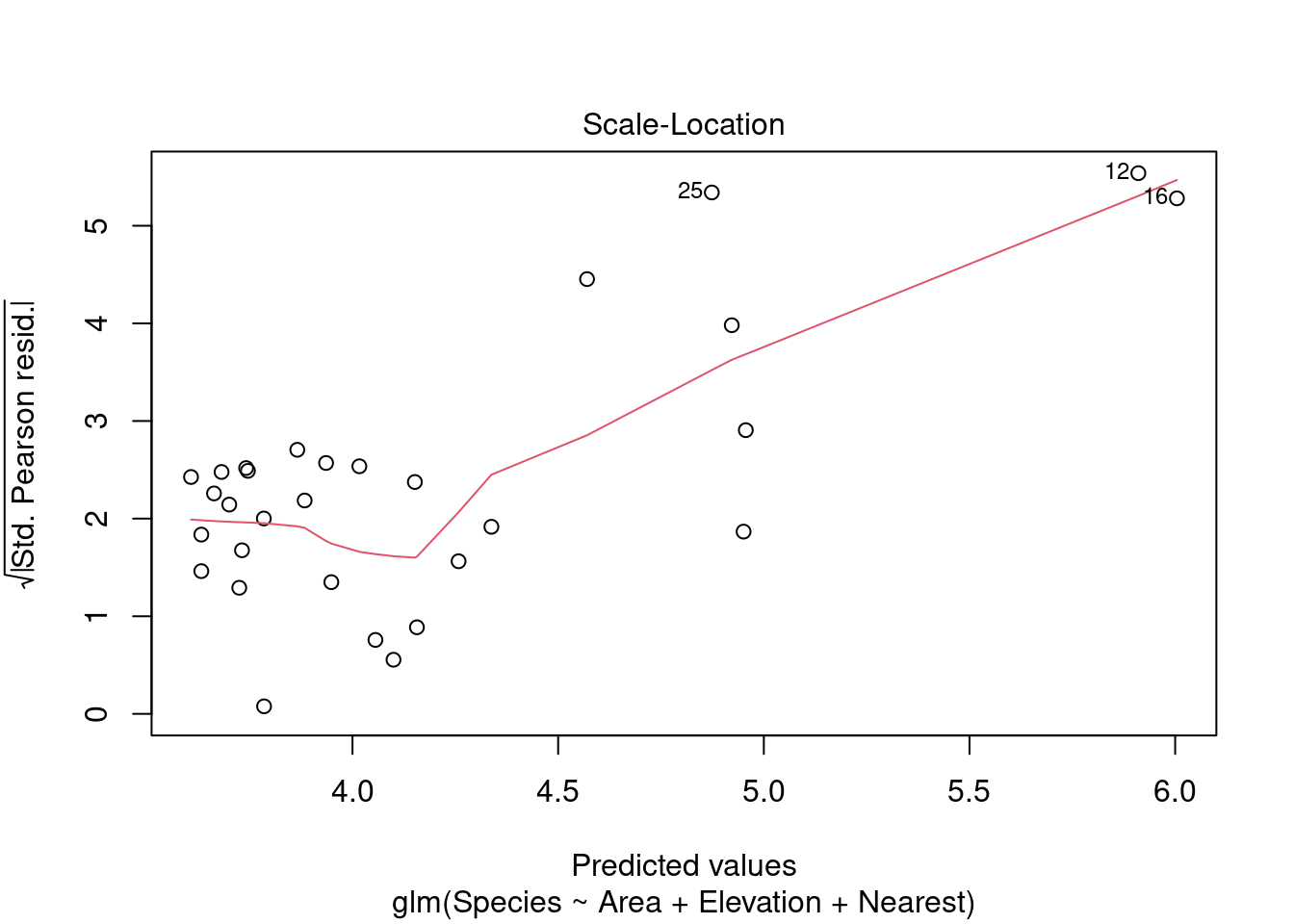
plot(modpl)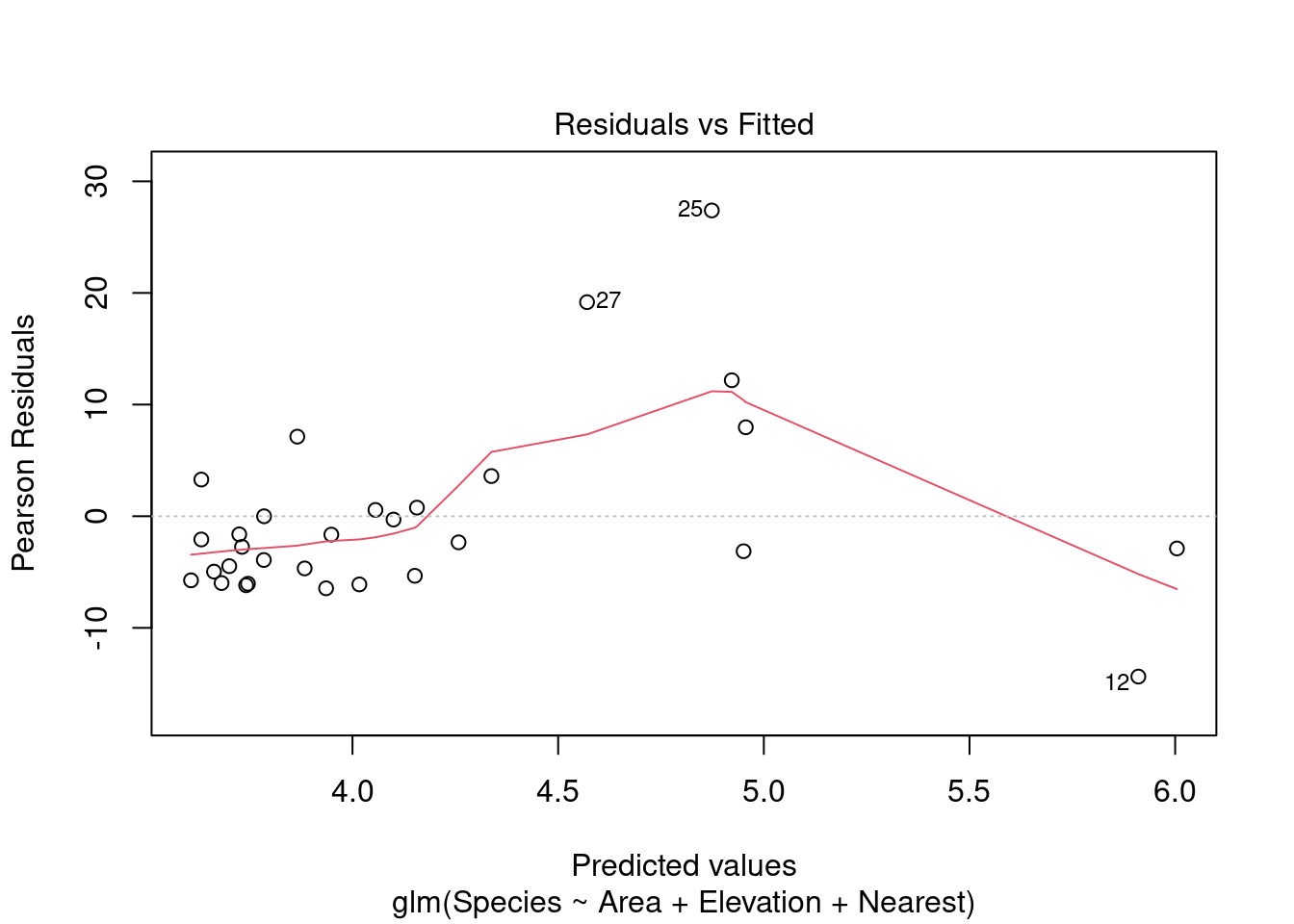
Warning in sqrt(crit * p * (1 - hh)/hh): NaNs produced
Warning in sqrt(crit * p * (1 - hh)/hh): NaNs produced